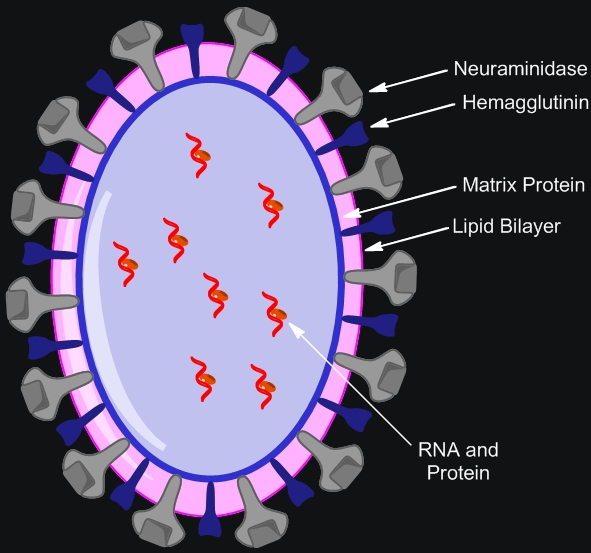
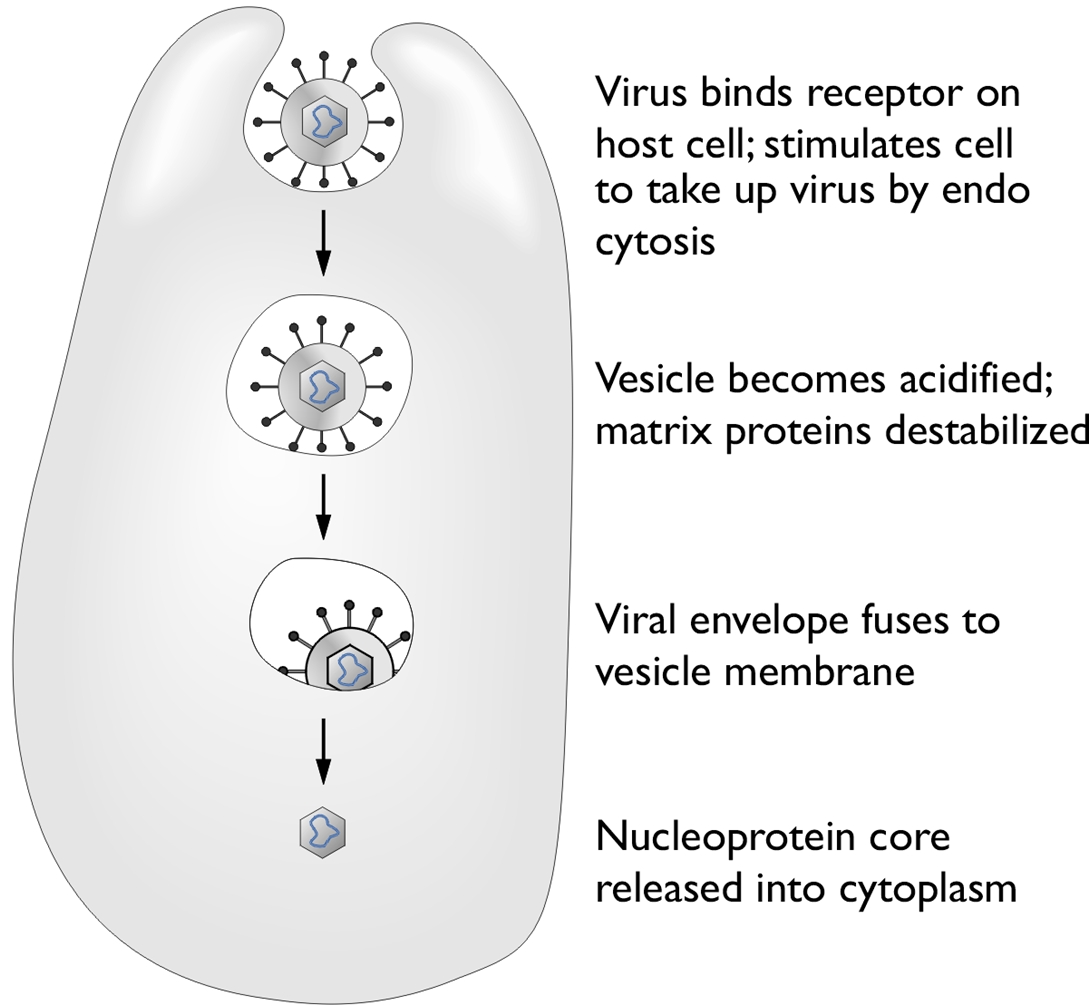
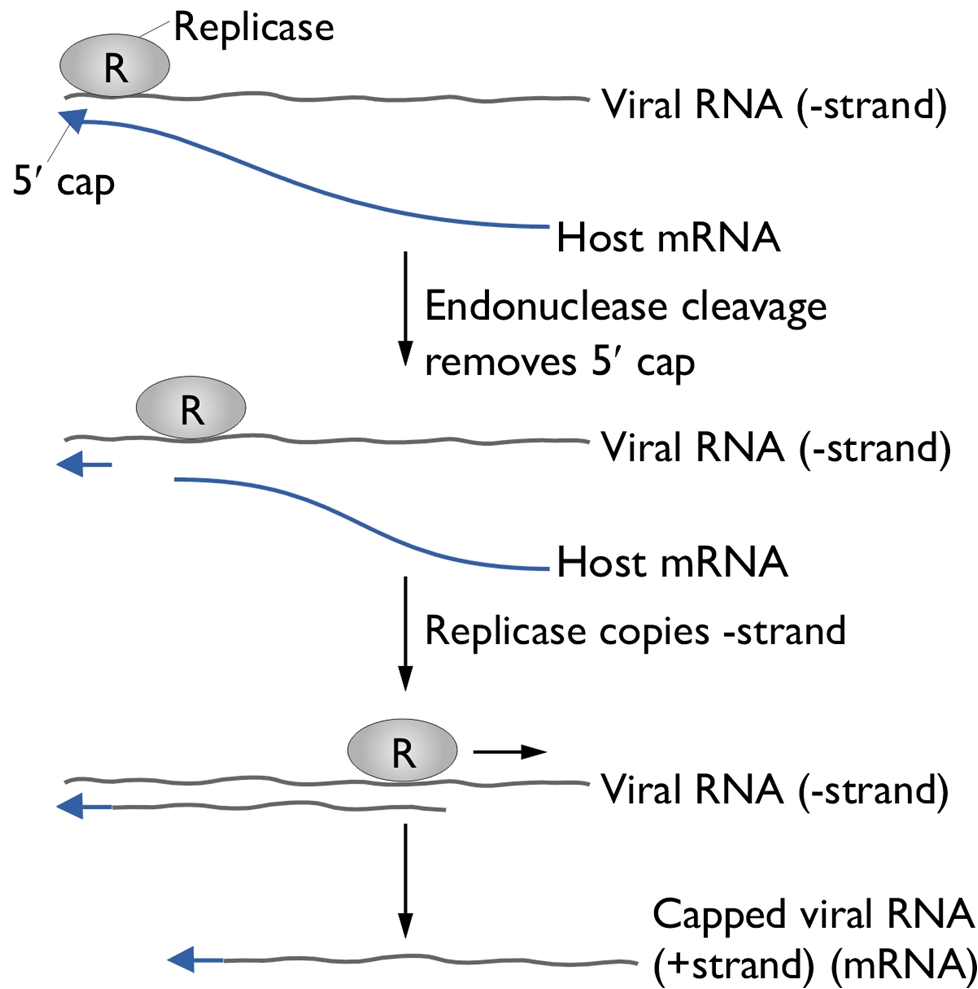
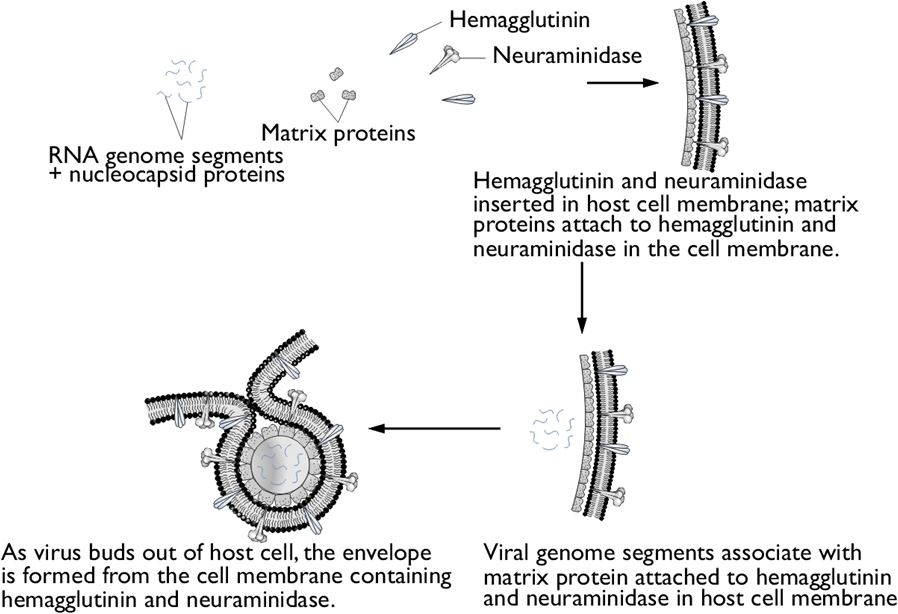
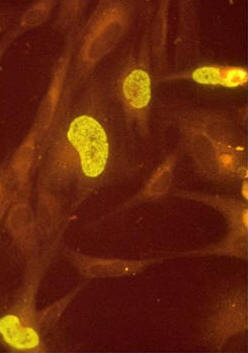
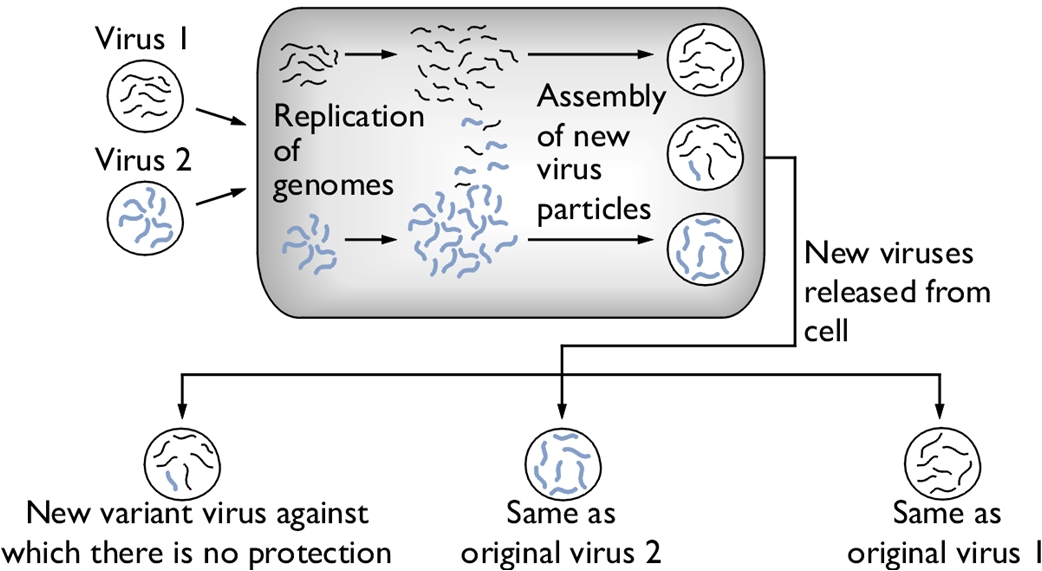
Influenza A VirusOverview: Influenza A virus is a single-stranded, negative-sense, enveloped RNA virus and a member of the Orthomyxoviridae family (Figure 1). This pathogen is spherical or globular in shape and roughly 100 nm (nanometres) in diameter; however, the virus typically displays a wide variation in size, ranging from as little as 8 nm to 200 nm. Influenza A virus infects upper respiratory mucosal membrane cells, causing an acute productive infection referred to as influenza. Figure 1. A photomicrograph depicting Influenza A virus. Pathogenicity: Targeted cells of the upper respiratory system are infected using hemagglutinin, a viral glycoprotein found on the virus' surface, that binds to sialic acid residues on host-cell surface glycoproteins and enters via receptor-mediated endocytosis (Figure 2). A low pH within the endosome or phagosome (engulfed vesicle) causes a subtle change in the structure of the viral hemagglutinin that enables it to fuse the viral membrane with the vesicle membrane, essentially releasing the viral genome into the cytoplasm (Figure 3). Thus, hemagglutinin is the protein which is central to both the receptor binding and fusion function of the virus. Figure 2. Schematic of the influenza type A virus. Click to enlarge. Figure 3. Entry & uncoating of Influenza A virus. The virus' genome enters the nucleus through the nuclear pores and is translated into viral protein. Since influenza A is a negative-sense RNA virus, it needs to convert its own negative-sense genome into a positive-sense strand so that it can use this newly formed positive-sense strand as a template to make many negative-sense RNA genome strands for progeny virion that are being generated. To do this, influenza virus subverts the host cell's translation machinery to its own, essentially stealing the 5' (five prime) cap of mature host cell mRNA in order to initiate the transcription of positive-sense RNA. The rest of the host mRNA is cleaved by endonucleases. Viral replicase then copies the negative-sense strand to produce a capped viral RNA (positive-strand) (Figure 4). The assembly of viral proteins and RNA creates new virions that exit the infected cell by budding off from the plasma membrane of the infected cell, allowing the spread of newly produced viruses to neighboring cells. Viral neuraminidase is an enzyme on the surface of influenza viruses that enables the virus to be released from the host cell. In other words, in order for the virus to be released from the cell, neuraminidase must enzymatically cleave the sialic acid groups from host glycoproteins as the virus is being exocytosed. Figure 4. Structure of the 5’ cap and the mechanism by which influenza virus subverts the host cell’s translation machinery to its own use. Influenza virus membrane contains two glycoproteins, namely, hemagglutinin and neuraminidase. While the hemagglutinin on the surface of the virion is needed for infection, its presence inhibits release of the particle after budding. Viral neuraminidase cleaves terminal sialic acid residues from glycan structures on the surface of the infected cell. This promotes the release of progeny viruses and the spread of the virus from the host cell to uninfected surrounding cells. Hemagglutinin and neuraminidase subtypes are used to classify different strains of the influenza type A virus and different combinations result in different host ranges and virulence. Budding of influenza virus through the host cell is illustrated in Figure 5. Figure 5. Budding of influenza virus through the host cell cytoplasmic membrane. Click to enlarge. Transmission and Identification: Birds are the main reservoir of the virus, and transmission usually occurs through contact with airborne droplets of bodily fluids from coughing or sneezing. Those over the age of 65 or those fighting a chronic illness are at the highest risk of infection and death; most mortalities are associated with a secondary infection with another pathogen. After an incubation period of 18 to 72 hours, infection can last seven to ten days followed by rapid recovery. Symptoms include: fever, fatigue, headache, muscle aches, sore throat, dry cough, and nasal congestion. Identifying Influenza type A can be difficult since many other pathogens initially cause similar symptoms. In order to identify the virus, viral cultures derived from mucous samples can be tested for positive hemadsorption, and the type can be determined using immunofluorescence (Figure 6). Type A and type B can also be distinguished by the number of proteins surrounding the RNA segments; for instance, type A has ten proteins, whereas type B has 11 proteins. Figure 6. Immunofluorescence staining of influenza virus-infected lung cells. The cells were treated with fluorescent dye-tagged antibodies against the specific virus. The nucleus of infected cells is bright yellow green, indicating the location of the viral antigen. Variants: Influenza A variants are produced by two mechanisms: antigenic drift and antigenic shift. Antigenic drift is the more common mechanism for generating variation. From year to year, point mutations accumulate in the genes encoding the viral surface hemagglutinin and neuraminidase antigens, potentially producing epitopes not recognized by preexisting antibodies. Thus, antigenic drift underlies the need for yearly influenza vaccination. To produce a new supply of influenza vaccines each year, manufacturers need to identify at least a year in advance (to account for the long time required to produce such a large quantity of vaccine) which type of drift is likely to emerge in the following fall/winter flu season. Antigenic shift occurs when influenza viruses from different host species, such as swine (pigs) and humans, both infect another species such as migratory waterfowl (Figure 7). When two dissimilar viruses infect the same cell, the viral nucleic acids, which consist of eight single-stranded RNAs, can undergo a reassortment to produce hybrid viruses with major antigenic changes. These new variants often cause severe infection in chickens, which subsequently can be transferred over to humans. Thus, to control the emergence of the new strain, all infected chickens are prompted to be eliminated. Figure 7. Antigenic shift in influenza virus. Click to enlarge. References: Baignent, S.J., & McCauley, J.W. (2003). Influenza type A in humans, mammals, and birds: determinants of virus virulence , host-range and interspecies transmission. BioEssays, 25: 657-671. |