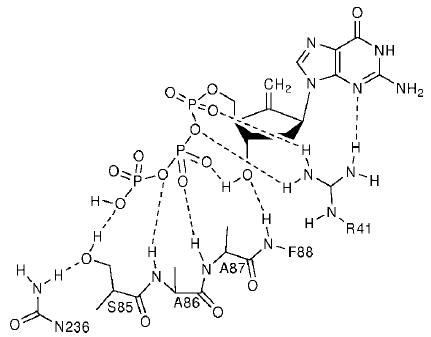
Hepatitis B VirusContents Mechanism of Action: Once entecavir enters the cell, cellular kinases phosphorylate it into an active triphosphate compound. Once active, the chemical compound inhibits the only known enzymatic target of the hepatitis B virus, hepatitis B viral polymerase. The mRNA encoding hepatitis B viral polymerase serves as the template for synthesis of genomic virion DNA through reverse transcriptase activity. The primer for viral polymerase DNA synthesis is a hydroxyl group of a tyrosine residue near the amino terminus of viral polymerase. While normal cellular polymerases and reverse transcriptase enzymes use free hydroxyl groups of DNA or RNA as primers to initiate DNA synthesis, hepatitis B viral polymerase depends on hydroxyl groups of amino acid residues, resulting in covalent attachment of viral polymerase to the progeny genome it produces. This priming is also unique in that the first three or four bases are template directed, using a stem-loop structure within the mRNA encoding viral polymerase. The resulting primer subsequently translocates to another portion of the genome to initiate full-length first-strand DNA synthesis. The entire polymerase activity occurs within a cytoplasmic nucleocapsid particle assembled from hepatitis B virus core protein, into which viral polymerase directs the inclusion of itself and its template. The final product is a partially single-stranded, partially double-stranded gapped DNA which is released in mature virions and repaired after translocation to the nuclei of newly infected cells (Langley et al., 2007). Entecavir triphosphate displays activity against the synthetic activities of hepatitis B viral polymerase by disrupting (1) the unique protein-linked priming activity, (2) RNA-directed first-strand synthesis via reverse transcription, and (3) second-strand DNA-directed DNA synthesis. Recall that the genome of hepatitis B virus is partially single-stranded (minus-sense) and partially double-stranded (plus-sense). In order to understand the interaction between entecavir and hepatitis B virus at the molecular level, entecavir triphosphate must be modeled in the catalytic site of hepatitis B virus reverse transcriptase, as well as in the process of hepatitis B virus DNA elongation. Unlike conventional hepatitis B viral treatments, entecavir is unique in having a D-configured cyclopentyl group. During translation of viral DNA, entecavir fits directly into the hydrophobic pocket of hepatitis B virus reverse transcriptase at the back deoxyribonucleotide triphosphate (dNTP) binding site via hydrogen bonds (Figure 6). Normal deoxyguanosine triphosphate (dGTP) cannot wholly occupy the pocket during replication since it does not contain a cyclopentyl group like entecavir (Figure 7). Recall that the chemical compound is a guanosine nucleoside analogue. This property accounts for the higher affinity of entecavir for the hepatitis B virus reverse transcriptase than that of dGTP, and higher potency compared to other drugs. The absence of this small pocket in human polymerase correlates with the relative inactivity of entecavir during host DNA replication (Langley et al., 2007).
Figure 6. Entecavir structure and hydrogen bonding network with hepatitis B reverse transcriptase.
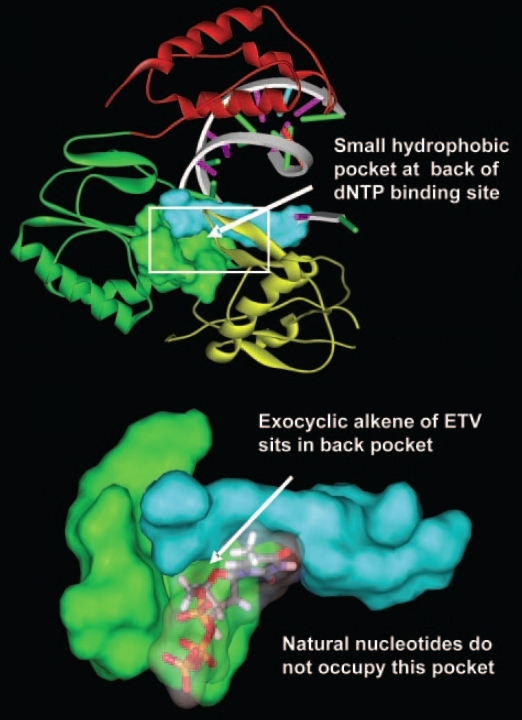
Figure 7. Homology model of hepatitis B virus reverse transcriptase reveals a novel hydrophobic pocket at the rear of the dNTP binding site (protein = green; DNA = cyan). Since entecavir possesses a potential 3’ (3 prime) hydroxyl group, it is important to establish the effect of entecavir incorporation on hepatitis B DNA chain extension. The mechanism for chain termination by this drug likely involves incorporation and abortive extension of entecavir-containing DNA. This can occur in any three instances, namely, (1) initial docking into the dNTP binding site and addition of entecavir onto the 3’ end of the growing DNA; (2) upon addition of ETV in the +1 position; and (3) upon further addition of nucleotides when entecavir is elongated to the +2 position. When entecavir enters hepatitis B virus reverse transcriptase, its cyclopentyl group causes steric strain on the hepatitis B virus DNA, thereby distorting and partially blocking the dNTP binding site. As a result, this prevents the binding of any new substrate and resulting in chain termination (Langley et al., 2007).
|